

JupyterLab is an interactive development environment for working with notebooks, code and data. It enables you to use text editors, terminals, data file viewers, and other custom components side by side with notebooks in a tabbed work area.
Main features:
What does research reproducibility mean? BY STEVEN N. GOODMAN, DANIELE FANELLI, JOHN P. A. IOANNIDIS SCIENCE TRANSLATIONAL MEDICINE01 JUN 2016 : 341PS12
"1,500 scientists lift the lid on reproducibility ": A survey article published in Nature, discussing whether the "reproducibility crisis" in scientific research is real, what the potential causes are and what we could do to make our research more reproducible.
"Challenges in irreproducible research": A special collection of Nature (July 2018), that addresses the growing alarm about results that cannot be reproduced.
"Scientific Rigour and Reproducibility": Another special collection of Nature (April 2018) that contain articles specifically targeted to discuss issues in research reproducibility.
"Reproducibility of research: Issues and proposed remedies": A special issue of PNAS (March 2018), focusing on issues and potential solutions for research reproducibility.
"Report on the First IEEE Workshop on The Future of Research Curation and Research Reproducibility": NSF report on reproducibility landscape, the importance of research curation, and calls out collaborative effort among publishers, research communities, and funders.
"Science Forum: Ten common statistical mistakes to watch out for when writing or reviewing a manuscript": A list of some of the most common statistical mistakes that appear in the scientific literature, as well as advice on how authors, reviewers and readers can identify and resolve these mistakes.
Best practices and good enough practices for computational reproducibility: "Ten Simple Rules for Reproducible Computational Research" and "Good enough practices in scientific computing"
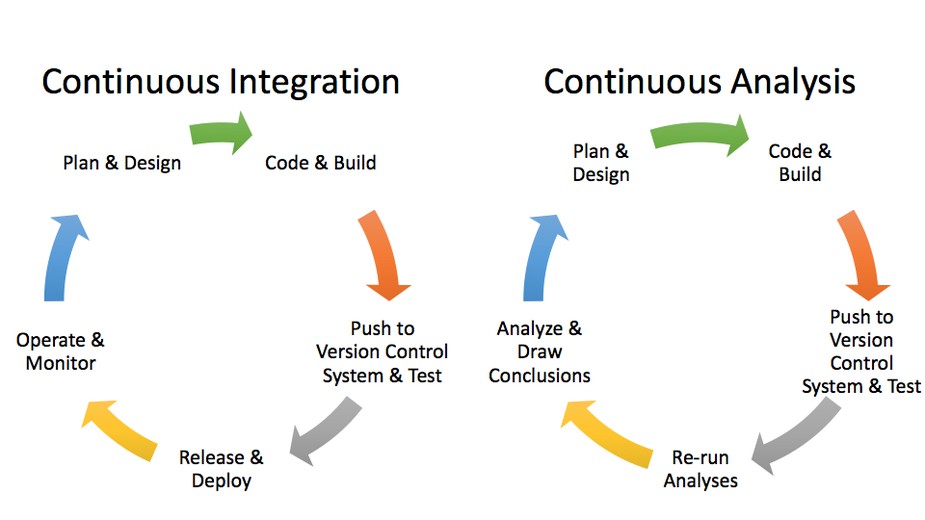
"Reproducibility: automated.": use “continuous analysis" workflow to automate and containerize data analysis steps, allowing others to easily reproduce and build on the results.

Collect articles and PDF from web browsers as you discover them
Organize, read and annotate in one place
Share with your group
Prepare references for Microsoft Word or LaTeX (BibTeX) with ease
Access your experimental needs
Institutional support (We are in the process of evaluating ELNs and purchasing a license. Feedback welcomed!)